Enhancing TCR-pMHC Binding Predictions
3A Internship Project Overview (DTU)
- Location: Technical University of Denmark (DTU)
- Group: Immunoinformatics and Machine Learning Group, led by Prof. Morten Nielsen
Description
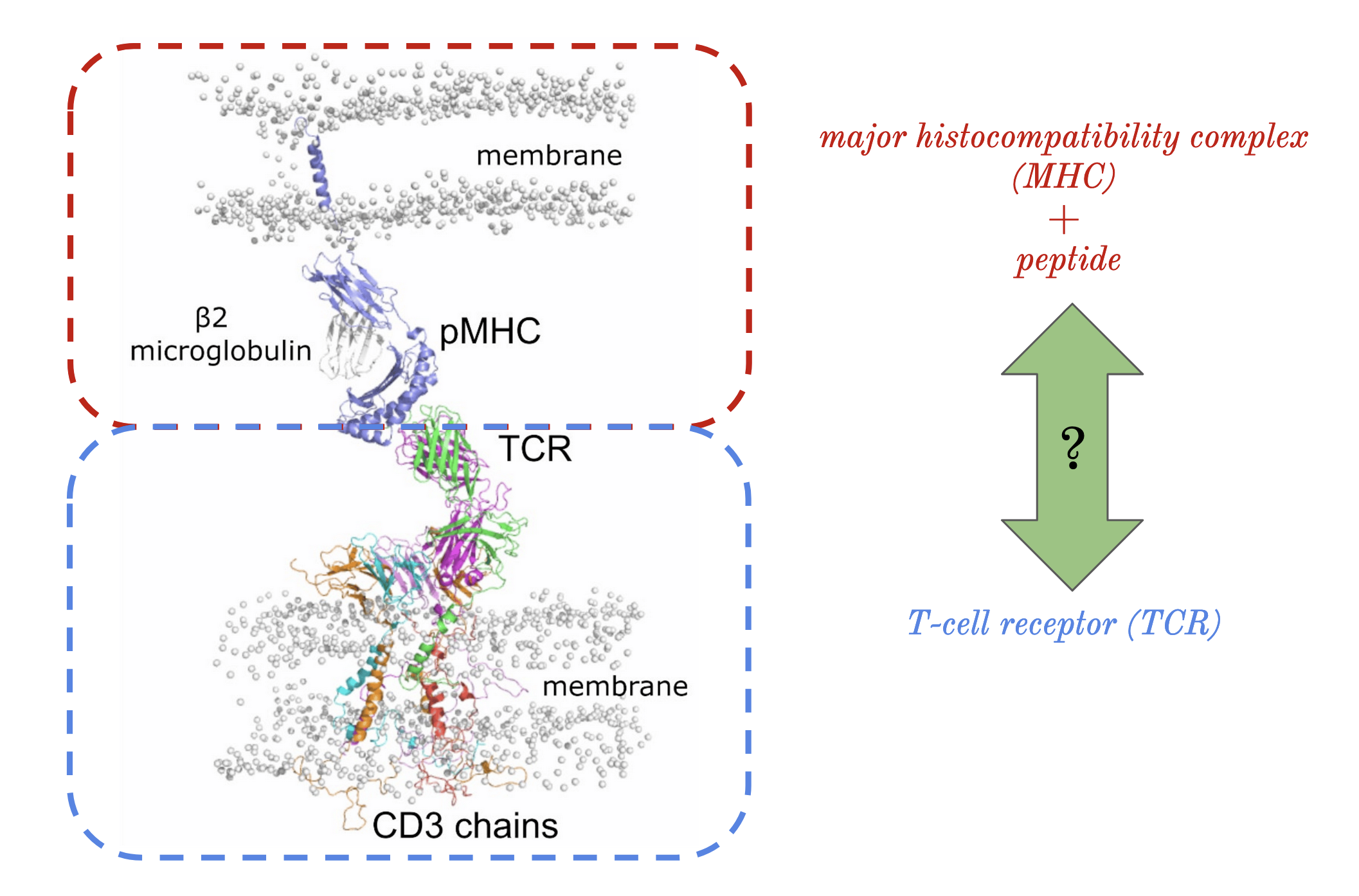
This project is dedicated to advancing predictive models for binding interactions between T-cell receptors (TCRs) and peptides presented by Major Histocompatibility Complex (MHC) class I molecules, with applications in vaccine development, cancer immunotherapy, and autoimmune disease management.
Research Goals and Innovations
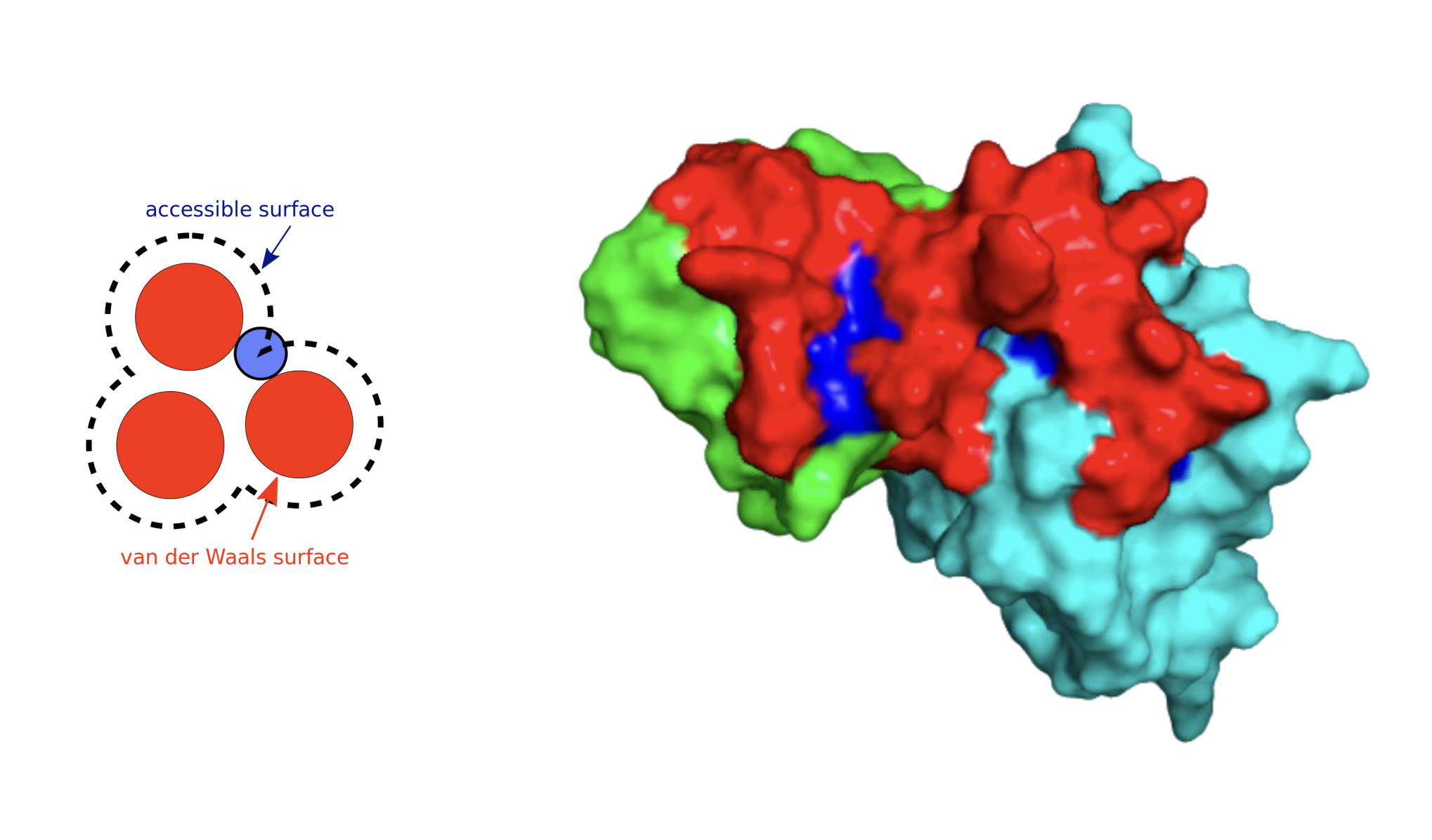
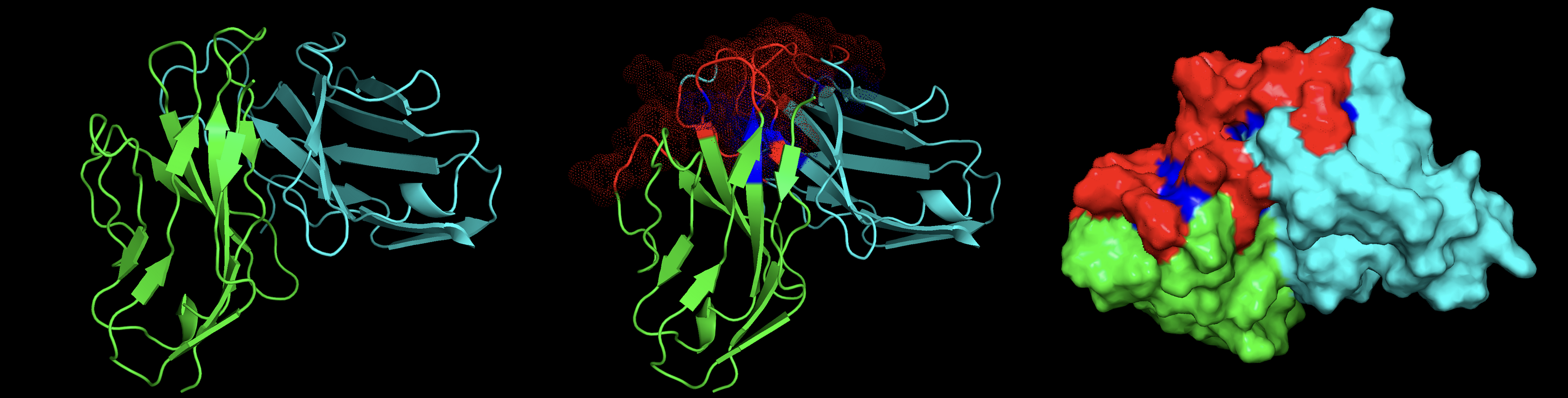
The central objective of my research was to develop a more accurate machine learning model to predict how TCRs recognize peptides displayed by Major Histocompatibility Complex (MHC) class I molecules. By refining current methods, my work sought to bridge gaps in prediction accuracy for peptides with limited known TCR interactions, a significant challenge in immunoinformatics. Key steps included using the RSA-based Shrake-Rupley algorithm to identify and prioritize solvent-exposed amino acid residues on TCRs, which are most likely to interact with peptides. Additionally, I employed ImmuneBuilder to rapidly and accurately predict TCR structures, allowing for efficient computational analysis.
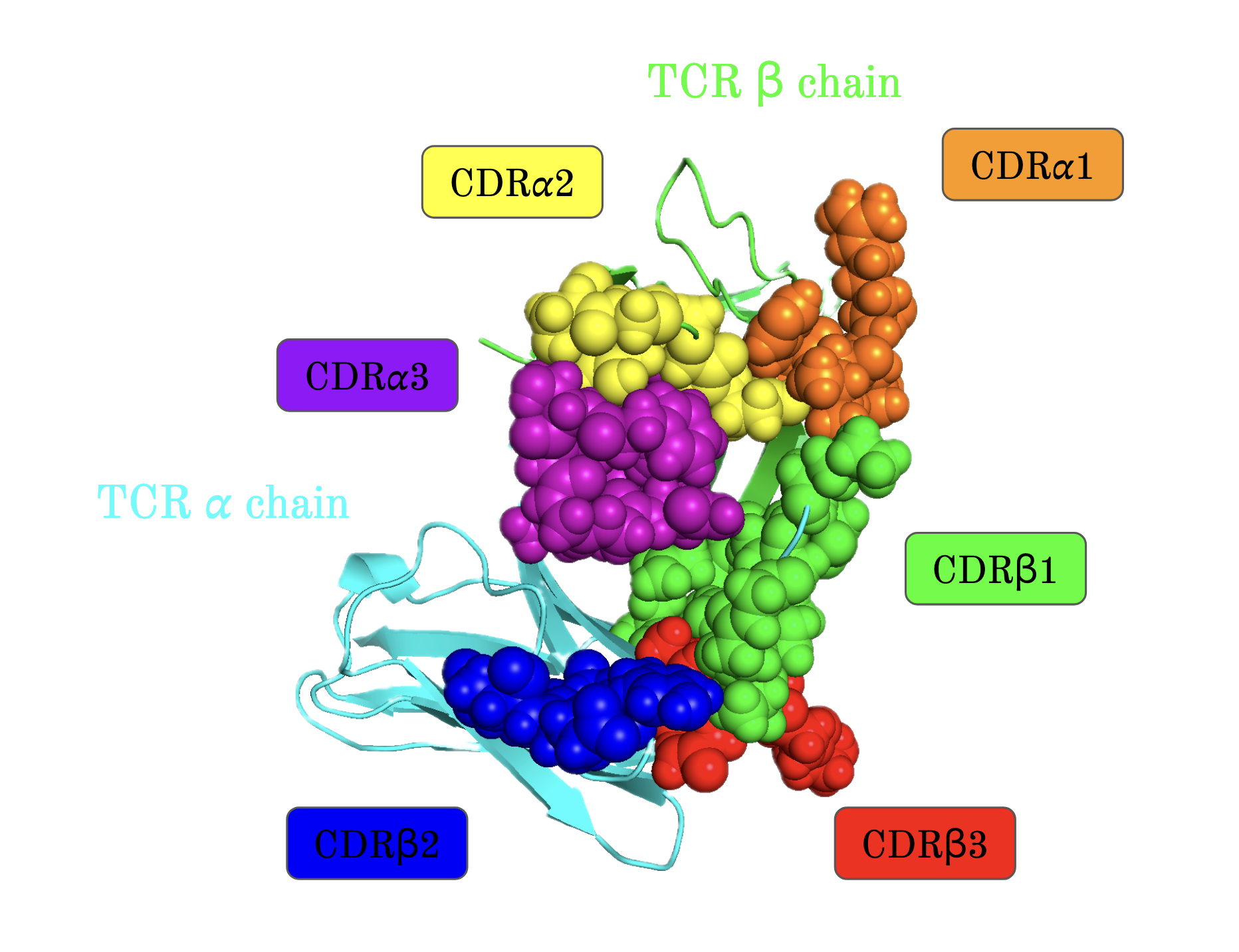
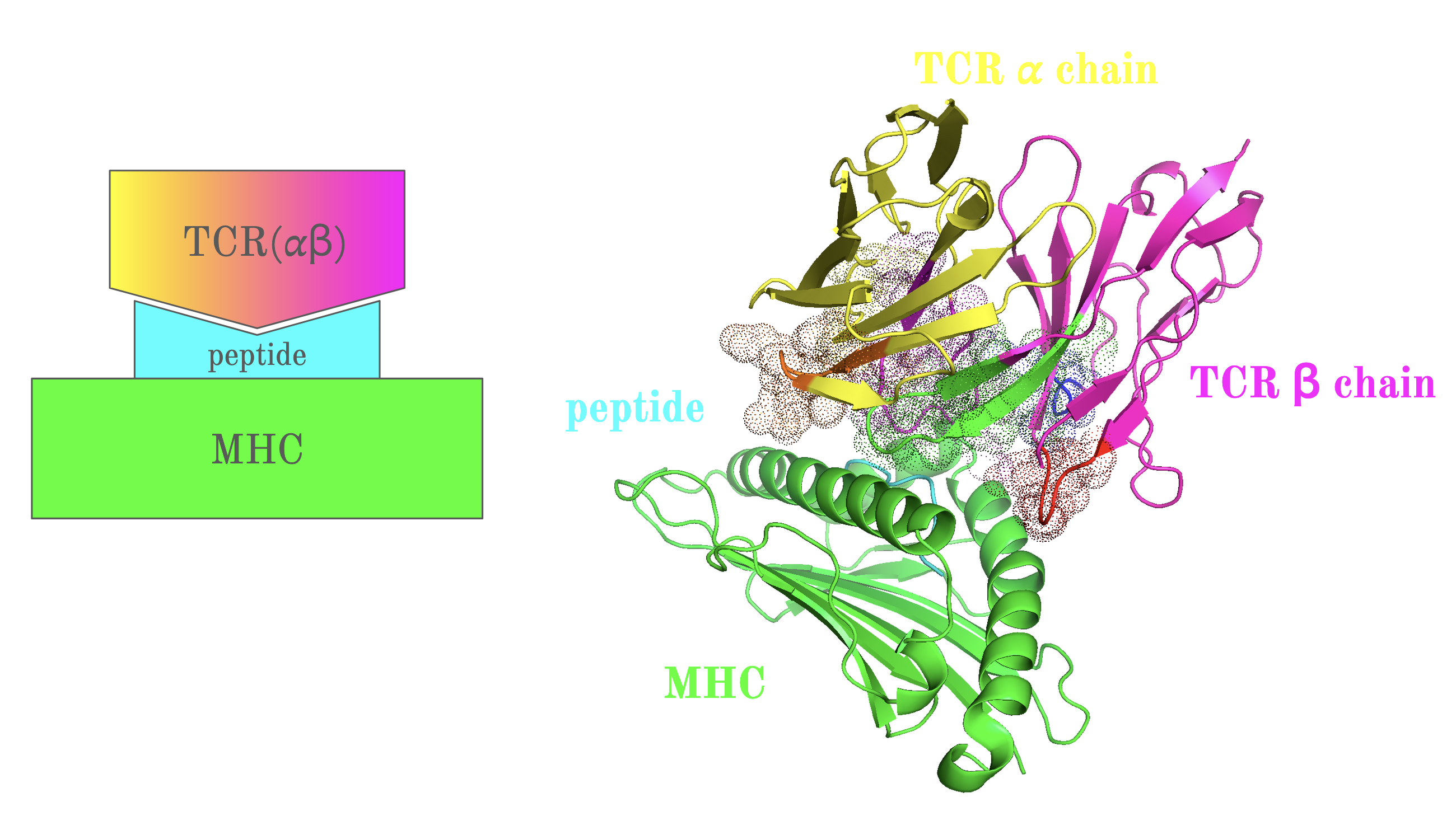
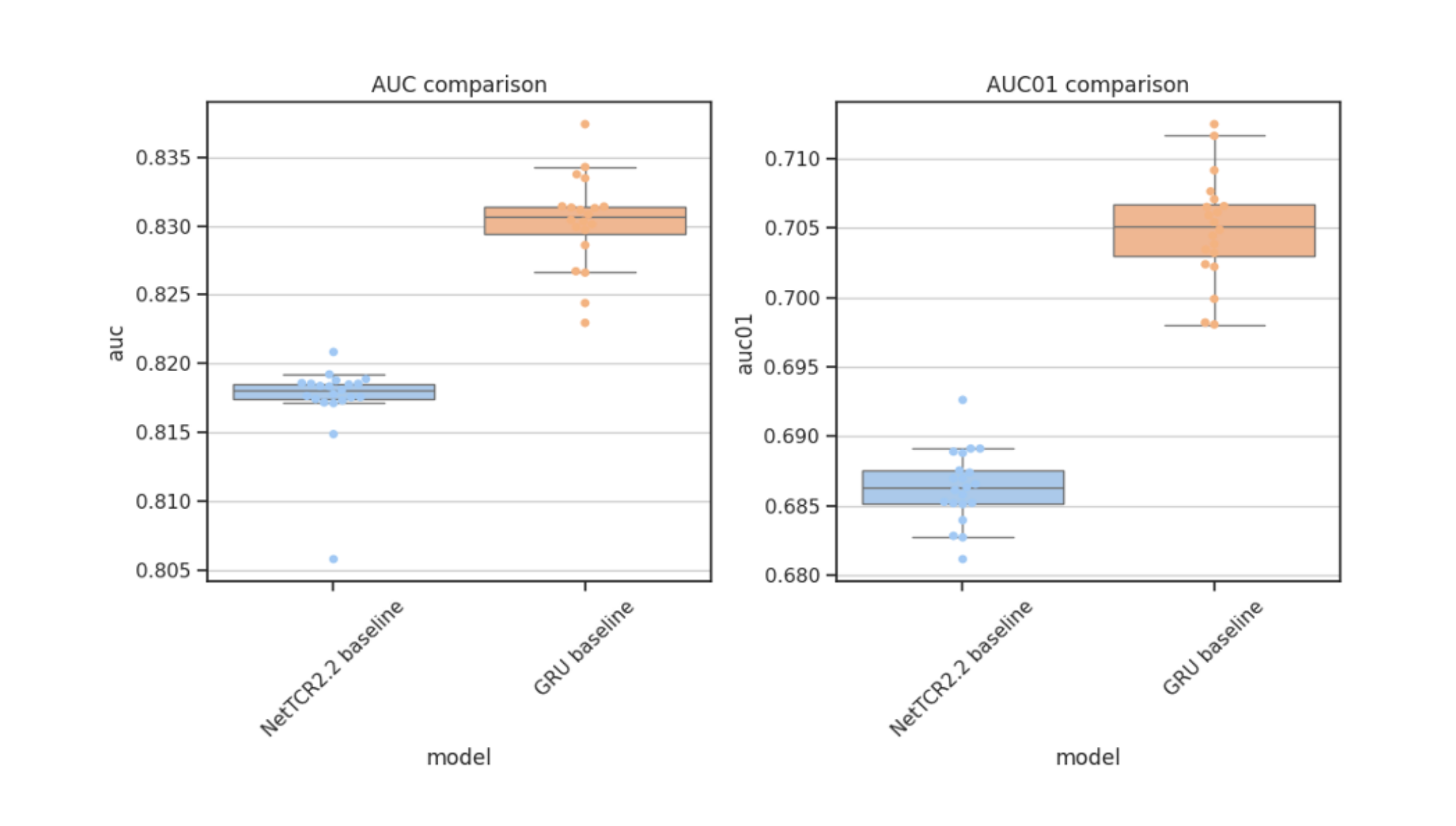
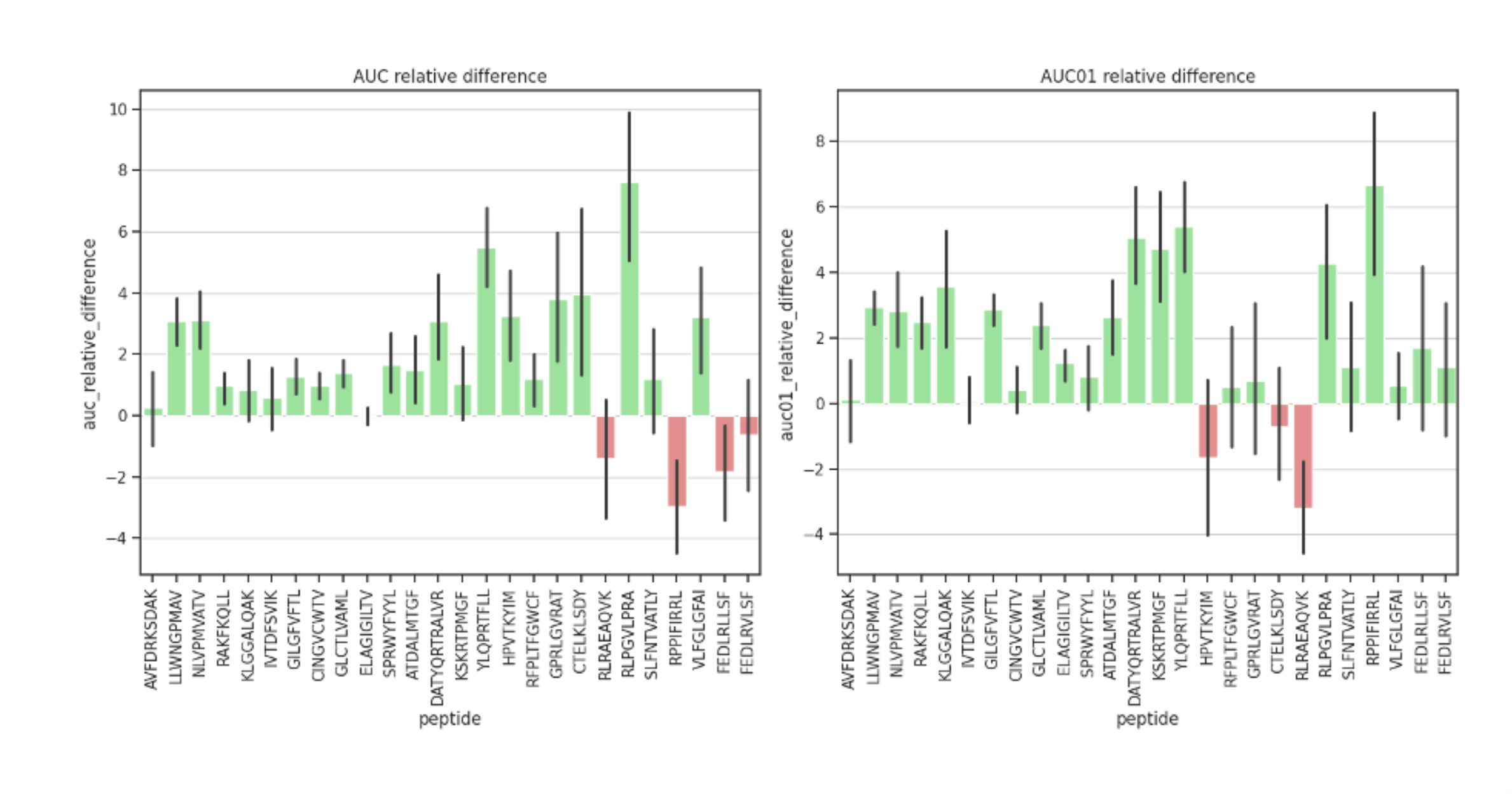
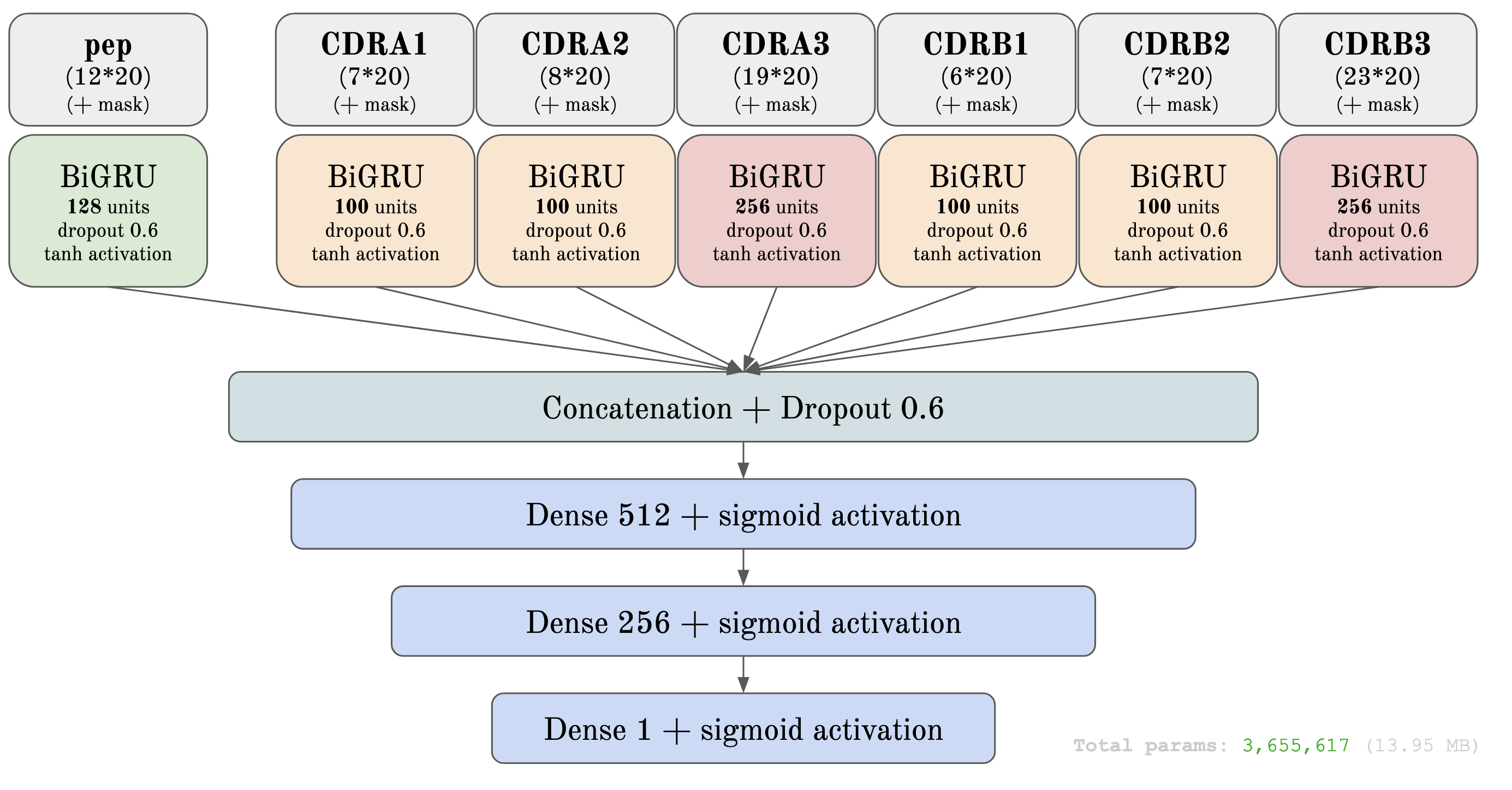
To address predictive limitations, I developed a novel machine learning model based on a bidirectional Recurrent Neural Network (RNN) with Gated Recurrent Units (GRUs). This approach enabled the model to capture the dynamic sequence patterns and structural features unique to TCRs and peptides, resulting in an improvement in prediction accuracy, specifically for peptides with fewer positive TCR interactions. Through rigorous validation, including nested cross-validation with early stopping techniques, the model demonstrated a 2.7% increase in AUC0.1, confirming its effectiveness over previous models.
An unexpected finding during this project was that RSA-based removal of residues, though theoretically sound, eventually reduced predictive performance. Consequently, I pivoted to using RSA as an additional feature rather than a filter, but it did not improved the precision either. Thus I only kept the new architecture without intergrating structural features.
Key Findings
- Feature Integration: Shifted from RSA-based residue removal to incorporating RSA as an additional model feature, preserving essential structural information.
- New architecture based on GRU layers instead of only convolutions
- Performance Improvement: Achieved a 2.7% increase in AUC0.1, validated through statistical tests, confirming significant improvement over previous models.
- Model Robustness: Enhanced accuracy in identifying peptides with fewer positive binders, demonstrating reliability across diverse peptide data.
Conclusion
Initially, our RSA-based approach aimed to retain only accessible residues essential for interactions, but filtering residues by RSA thresholds unexpectedly lowered model performance, likely by excluding valuable structural information. Recognizing RSA’s value, we adjusted our strategy to integrate it into the model without removing residues. Building on previous CNN models, we developed a GRU architecture tailored to capture sequential dependencies in TCR-peptide interactions, incorporating structural features like RSA and minimum distance values. Although these features enriched the model’s understanding, they did not yield the expected gains in accuracy, highlighting the complexity of integrating structural data. Rigorous testing showed the GRU model improved predictive performance, especially with challenging peptides, yet RSA and distance features did not consistently enhance results, indicating further exploration is needed for effective feature integration.